Cases Studies
Case studies are used to test and evaluate the solutions provided by OpenRiskNet to the predictive toxicology and risk assessment community especially regarding the usability of the developed APIs and the interoperability layer. These are demonstrating the capabilities to satisfy the requirements of the different stakeholder groups including researchers, risk assessors and regulators and present real-world applications like systems biology approaches for grouping compounds, read-across applications using chemical and biological similarity, and identifying areas of concern based on in vitro and in silico approaches for compounds lacking any previous knowledge from animal experiments (ab initio case). The case studies defined below are guiding the prioritisation of data sources and tools to be integrated and used as first examples to improve the level of the corresponding APIs with respect to harmonisation of the API endpoints, service description and semantic annotation.
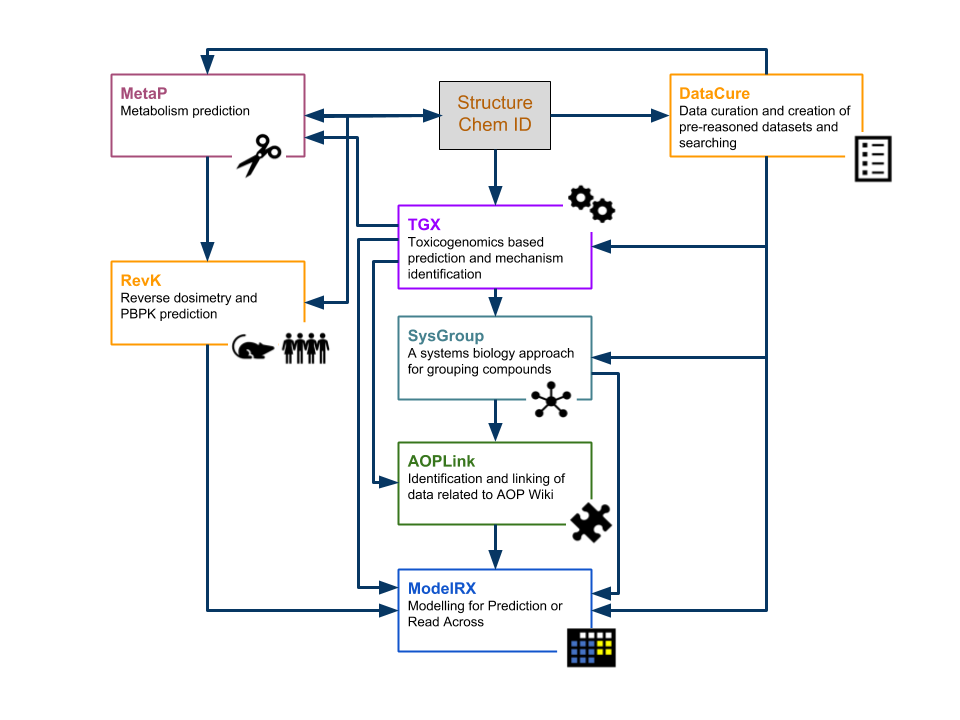
| DataCure | Data curation and creation of pre-reasoned datasets and searching |
| ModelRX | Modelling for Prediction or Read Across |
| SysGroup | A systems biology approach for grouping compounds |
| MetaP | Metabolism Prediction |
| AOPLink | Identification and Linking of Data related to AOPWiki |
| TGX | Toxicogenomics-based prediction and mechanism identification |
| RevK | Reverse dosimetry and PBPK prediction |
A workflow for the safety assessment of chemicals without animal testing developed within the SEURAT-1 initiative and was selected to guide the initial definition of the case studies. It aims to implement an approach to chemical safety assessment without relying on any animal testing; instead, constructing a hypothesis based on existing data, computational modelling, biokinetic considerations and, then, by targeted non-animal testing (Berggren et al., 2017). For further developments, other concepts will be added or followed in order to avoid any limitations created by a single framework, and this information will be incorporated into the related case study description.
Use cases
Testing and validation of basic concepts considered within the case studies has to be done in parallel to the development, in order to identify any issues as soon as possible. To be able to do so, fully elaborated subparts of the case studies have been translated into use cases to evaluate important key components of the infrastructure. A description of the scientific goals pursued and the technical areas of the infrastructure probed by the use cases is given below.
UC1: Merge existing data by a common structure identifier
This use case describes the need to generate a consistent set of data by merging multiple individual datasets. An example would be generating activities for a set of structures against a particular enzyme target. The dataset generated could be the input for building a predictive model (see UC2). The assumption is that data for individual assays are accessible as datasets through an API (e.g. ChEMBL or PubChem web services) and that there is some mechanism for the user to select particular datasets from those that are available. Multiple datasets are merged into a single dataset based on identification of structures that are common between the datasets.
This use case is intended to indicate the capabilities of the infrastructure regarding data APIs. In the first step, the existing APIs will be evaluated regarding the search and download possibilities and how the data can then be transformed to get a harmonized view across the different data sources. The equality of the assays and the comparability of the data in the different databases has to be evaluated manually by the user but this will be more and more automated based on the semantic interoperability layer during later development steps of this use case.
UC2: Building and using a prediction model
Use case 2 describes the process of building predictive models from OpenRiskNet compliant datasets and applying the generated models on prediction datasets. Datasets used for training and prediction, can be the result of use case 1.
This use case can evaluate the progress of modeling APIs’ integration into the OpenRiskNet ecosystem. It can test the maturity of the semantic interoperability layer to efficiently distinguish if a training dataset is compatible with an algorithm and if a prediction dataset is compatible with a prediction model. Moreover, the modeling APIs are required to publish the generated models and datasets accompanied with semantic metadata on their life cycle, whenever requested. Thus, this use case will also be able to evaluate the process of semantically enriching the dynamically created entities.
Formal description →
Pseudocode →
UC3: Search and Retrieve Assay Data based on Ontological Terms
For Key Events (KEs) in Adverse Outcome Pathways (AOPs) to be useful in decision making, the event needs to be observable. That is, an experimental measurement needs to indicate that the key event happens and will trigger downstream events. Various other European projects are working on linking KEs to bioassays (EU-ToxRisk, etc.), but to explore this potential link, we need an infrastructure to allow researchers to find experimental data for some type of assay for some type of (nano)material. One aspect of that is to find omics data for a particular material.
Use Case 3 describes the basic process of finding and retrieving omics data related to a (nano)material and a specific KE. The KE is related to some bioassay, and using an ontology approach, e.g. with BioAssay ontology terms can be used to query databases with experimental omics data.
This use case can evaluate the progress of the ontology services as well as the coupling of ontological terms with regards to data and assays. Using additional resources that link AOP KEs to genes, proteins, or assays, such as those being set up on the WikiPathways AOP Portal, it generated a needed level of interoperability to support and possibly validate the European research for replacing animal testing with AOP-guided in vitro omics approaches.