Use case 1
Merge existing data by a common structure identifier
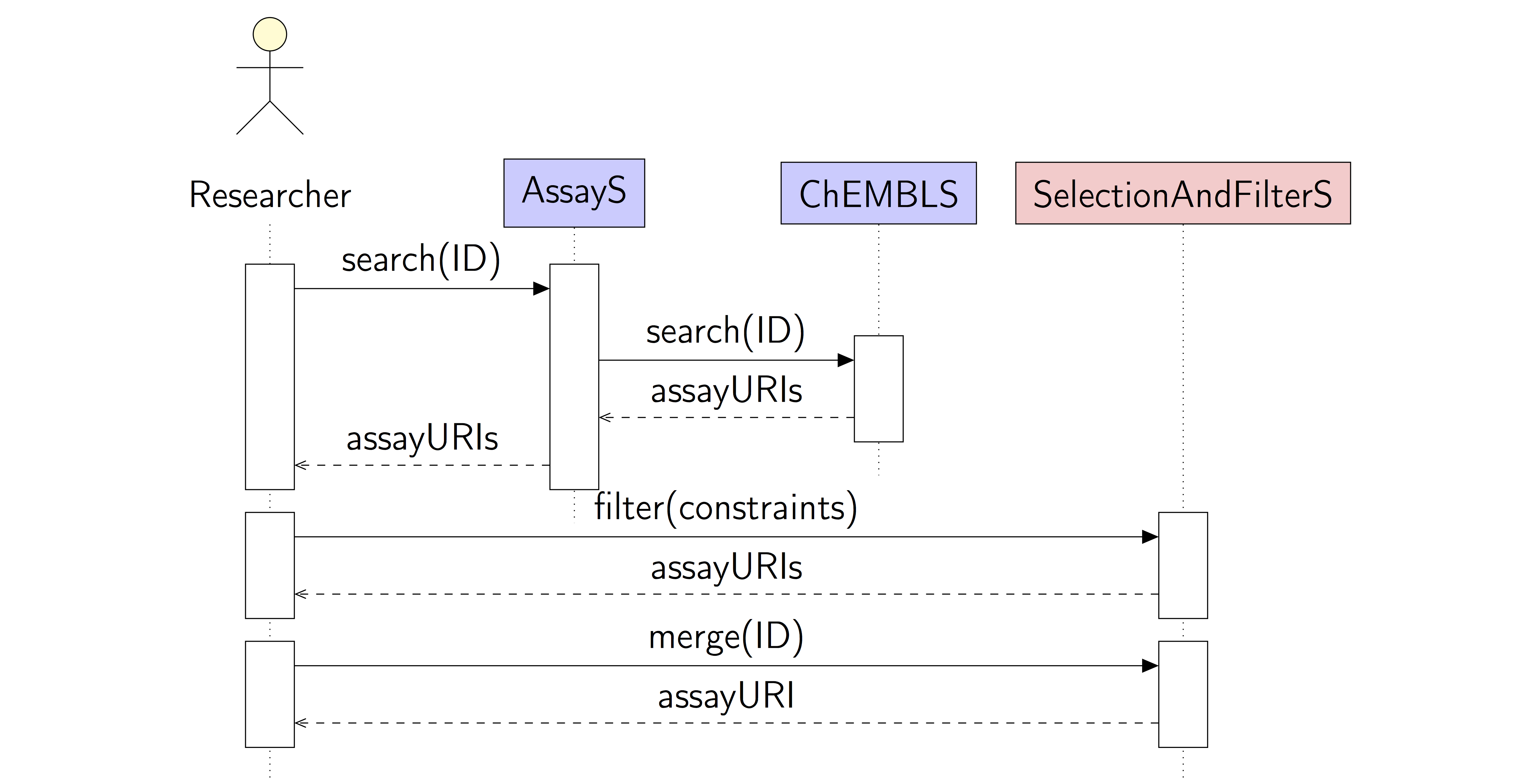
Diagram
Name:
Merge existing data by a common structure identifier
Brief Description:
A user searches for existing assay information, selects the desired information, and merges the results based on a unique structure identifier.
Actors:
A researcher with background in toxicology.
Basic Flow:
The user supplies an identifier for a protein target, e.g. by name, UniProt ID, or ChEMBL ID, and searches ChEMBL for assays. The search result might be filtered with a number of simple constraints, e.g. words occurring in the description. The user selects the relevant assays. The assays are merged based on the common ChEMBL structure ID and the IC50 values. The results (structures and activity values) are stored as a new dataset and returned to the user.
Preconditions:
- Valid search criteria for the assay of interest
- A de-duplication strategy
Postconditions:
Success End Condition:
- At least one assay has been merged and the information has been returned
Failure End Condition:
- No assay has been found according to the supplied criteria
- No assay has been found according to the additional constraints
Minimal Guarantee:
- None
Relationship to other use cases:
The output of this use case could be an input to UC2.
Alternate Flows:
In case of duplicates, a strategy is required to resolve these multiple occurrences. To avoid to have to supply that strategy manually or during the search process, this strategy should be supplied in advance. The suggested merge strategy in case duplicates are encountered might be as follows:
- One assay has priority over another, ie. the order of the merge becomes important
- Merge using mean values only when values are consistent (e.g. within 2-SD range)
- Provide mechanism to interconvert data when specified as different data types (e.g. IC50 and Ki) or in different units (e.g. micromolar and nanomolar).
- Present all values and let user decide which to keep. Record user’s choice.
Exception Flows:
Possible exceptions:
- No suitable assays are found: the system returns an error code
- The supplied parameters are out of scope: the system returns an error code
- Merging fails: the system returns an error code
- De-duplication fails: the system returns an error code
- Inconsistent de-duplication choices by the user: the system returns an error code
Extensions:
- Alternative assay providers such as PubChem can be used.
- Allow merging using canonical SMILES instead of ChEMBL structure identifier.
- The datasets retrieved contain different variants of the chemical structure (e.g. different salt forms). Before merging a standardized representation needs to be generated.
- Employ standardised units for merging using activity values
- Extend UC to be merged using categorical manner, i.e. active vs. inactive, mutagenic vs. non-mutagenic