The OpenRiskNet Approach towards a Semantic Interoperability Layer - Part One: Problem Definition for Datasets
Harmonization and interoperability between data and computational services, the facilitation of their easy discovery, the sharing and usage of data sources as well as of the processing, analysis and modelling tools and finally a better documentation and reproducibility of the risk assessment workflows are major challenges that OpenRiskNet tries to address. The approach adopted by us to tackle these challenges is the development of a semantic interoperability layer to organize the communication between the user and the services or between two services. The blog post describes the taken approach and the first steps to describe data sources in this layer.
Thomas Exner (Douglas Connect), Chris Evelo and Egon Willighagen (Universiteit Maastricht), Marc Jacobs (Fraunhofer Gesellschaft zur Förderung der Angewandten Forschung e.V.), Philip Doganis and Haralambos Sarimveis (National Technical University of Athens), Iseult Lynch (University of Birmingham), Stefan Kramer (Johannes Gutenberg Universität Mainz), Cedric Notredame (Fundacio Centre de Regulacio Genomica), Danyel Jennen (Universiteit Maastricht), Georgios Gkoutos (University of Birmingham), Ola Spjuth (Uppsala Universitet), Paul Jennings (Vrije Universiteit Amsterdam), Tim Dudgeon (Informatics Matters), Frederic Bois (INERIS) and Barry Hardy (Douglas Connect).
The “OpenRiskNet: Open e-Infrastructure to Support Data Sharing, Knowledge Integration and in silicoAnalysis and Modelling in Risk Assessment” project, funded by the European Commission within Horizon2020 Programme, intends to improve the harmonization and interoperability (i.e. the ability to work together, be combined to workflows and transfer data between each other without manual intervention) of data and software tools used in predictive toxicology and risk assessment. This includes (1) in vivo, in vitro and in silico data and derived knowledge sources, (2) the processing, analysis, modeling and visualization services, in order to facilitate their easy discovery, sharing and usage. The vision is that knowledge management, including a better documentation and reproducibility, will lead finally to validated risk assessment approaches for safe-by-design studies and regulatory setting supporting the goals of replacement, reduction and refinement (3R). The approach taken by OpenRiskNet to tackle these challenges is based on the development of a semantic interoperability layer responsible for the communication between the user and the services or between two services. This semantic layer will provide detailed annotations on (1) the scientific background and the meaningful usage of the services and their limitations, (2) the required input as well as (3) the obtainable results including possible output formats and standards used. In order to generate the technical solutions for this layer, some domain specific standards have first to be developed to describe the data sources and computer services in this metadata-rich fashion. This first part of a multi-part post is intended to provide background information on the current status of data sharing efforts and guidelines enforced by funding agencies and publishers’ and describes the problems we are additionally facing with respect to the interoperability layer. Additional posts will follow whenever we have at least partly solved a problem.
FAIR Principles:
The most important resource for an empirical science like toxicology and its application in risk assessment is data, since it is the basis for collecting (weight-of) evidence and generating knowledge. Unfortunately, data is very often not valued enough and is only seen as a necessary step on the way to a scientific publication. Sharing and reusing data was, and is still not, seen as important or even needed for advancing personal careers; indeed closed data is even seen as a competitive advantage by some. However, data used only once and then locked up on local storage is strongly contributing to the irreproducibility crisis and is a waste of resources since the experiments or computations have to be done over and over again. Therefore, funding agencies and publishers push towards open data. For example, new projects funded under the Horizon 2020 program of the European Commission have to provide data management plans (for more information see the Guidelines to the Rules on Open Access to Scientific Publications and Open Access to Research Data in Horizon 2020 and How to create a DMP Plan) describing how the project will implement the FAIR principles (Findable, Accessible, Interoperable and Reusable, Mark D. Wilkinson et al.), which are reproduced here from the FORCE11 website with some additions resulting from experiences in the ELIXIR project marked in bold:
To be Findable, a dataset requires that:
Findable should in fact be both about the resource (using harmonized descriptions according to BioSchemas, DATS/DCAT or possibly Beacon-like solutions) and the actual metadata for the contents.
F1 (meta)data are assigned a globally unique and eternally persistent identifier.
F2 data are described with rich metadata.
F3 (meta)data are registered or indexed in a searchable resource.
F4 metadata specify the data identifier.
To be Accessible, a dataset requires that:
A1 (meta)data are retrievable by their identifier using a standardized communications protocol.
A1.1 the protocol is open, free, and universally implementable.
A1.2 the protocol allows for an authentication and authorization procedure, where necessary.
A2 metadata are accessible, even when the data are no longer or not publicly available.
A3 metadata should provide access protocols or show licenses (often also mentioned in the context of re-use).
To be Interoperable, a dataset requires that:
I1 (meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation.
I2 (meta)data use vocabularies that follow FAIR principles.
I3 (meta)data include qualified references to other (meta)data.
I4 when it is clear what the data should be interoperable with, tools should be available to link between these data standard formats, vocabularies, IDs,…
To be Re-usable:
R1 meta(data) have a plurality of accurate and relevant attributes.
R1.1. (meta)data are released with a clear and accessible data usage license.
R1.2. (meta)data are associated with their provenance.
R1.3. (meta)data meet domain-relevant community standards.
R2 (meta)data should be rich enough to allow its re-use in new applications not limited to only reproduction of the original results.
The FAIR principles are one big step in the right directions since they force the researcher producing data to upload the data to a public database, which provides the technical solutions for a globally unique and eternally persistent identifier as well as searching and browsing functionality for the metadata. On the other hand, they don’t provide guidelines on the format of the data or the information to be covered in the metadata, either for indexing in the database or for describing the experimental setup even if it is clear that this metadata is essential for understanding the data and, therefore, for its re-usability. It is clear that such domain specific standards cannot be covered by the general guidelines in the FAIR principles. However data organized in an Excel sheet or a CSV file in a user defined format also comply with the FAIR principles since the access protocols are standardized and universally implemented. However, reusing data structured in different ways is very hard and the interoperability can only be provided by analysing the format of each individual dataset and manual harmonization to align with other datasets. Community-wide, and where possible cross-community-wide, standardization is therefore essential for effective data sharing. While data formats are often defined by platform providers (e.g. cel files) and are accepted as standard, initiatives to standardize metadata files like ToxML and SEND fight with acceptance issues especially in academic settings when they are not enforced by regulatory agencies. One valid argument is that they are not flexible enough and are not updated quickly enough to keep pace with the large amount of emerging new techniques and the improvements of existing ones requiring adaptations of the data and metadata to cover all experiment-specific parameters.
Semantic Annotation to Reduce Standardization Requirements:
For reusing data it is important that all data and metadata for a full documentation of the experiment and relevant for re-use in new applications (see principle R2 above) is available and that it can easily be understood. It is clear that not all possible applications can be foreseen and some might need additional information, which seem to be irrelevant at the time of data generation. However, this just highlights that the approach of minimal datasets should be avoided and especially metadata formats should be extendable. In contrast, the specific format is not that important if information is provided on how to restructure the data according to the needs of the use case. This does not mean that we should not use standard file formats if they exist. For example, cel files for transcriptomics data and sdf for molecular structures are widely accepted and supported in many software tools. However they have still to be accompanied by meta(data) with a less well defined format but which still has to be uniquely identifiable.
1) Data and Metadata Guidelines
The OpenRiskNet project is committed to best practices of data management (demonstrated in previous projects like diXa and ToxBank) and will further develop them for the fields of toxicology, safety and risk assessment. Since data quality is influenced by procedures adopted during data collection, storage and treatment, detailed documentation including the relevant metadata on how data is collected (metadata related to the experimental procedures including references to more detailed protocols and standard operating procedures) and treated (description of manual and automatic manipulation, which in the latter case would include executables, scripts and/or workflows of the procedures) provides the necessary means to evaluate the quality of the resulting dataset. The combined availability of the metadata with the data is essential for further processing and analysis of the data, as well as for model development and evidence extraction based on this data. In this way, experimental conditions influencing the results can be identified as reasons for irreproducibility and can be integrated into modelling approaches to allow for comparison / integration of data from different sources with slightly varying experimental protocols.
To allow for the data re-use without the need to derive them from external sources like publications, final reports, working papers or lab books, all important information should be included in the data submission. Note that such descriptions still need to be referenced to understand the background of a specific study, for provenance and for correct credit. In a paper of Robinson et al., the problems of how to define and evaluate data completeness and quality are addressed in great detail. Information for toxicological datasets should include descriptions of the tested chemicals, exposure scenario, test system and data collection methods as well as results (raw and processed). The description of data collection should include the data collection process, instruments used (including scale, resolution and temporal coverage), hardware and software used, and secondary data sources used. Even if most of the time only minimal, predefined information are required for data upload to standard data repositories, we would encourage a general motion of “the more the better” since some experimental parameters might become important when datasets are combined to collect information from a broader set of molecules e.g. for generating a predictive model.
Of course the drawback of “more” is that this also involves more work and that standards for the description of this extra information are often not yet available. Community standards that describe and standardise expected useful information are much needed. Approaches that automate and thereby facilitate the upload of the descriptive data (for instance through the development of eNotebook data standards with automated data upload to repositories for completed data sets) and approaches that make the collection of such extra data more directly useful (e.g. tools that perform power calculations automatically when sufficient study descriptions are provided or that import additional information about food composition automatically) should be developed and integrated with the data repository. Such dataset enrichments could be triggered automatically during data upload or manually and should be clearly marked and stored as derived data with additional annotations. In a similar way, manual annotations and curations (e.g. outlier identification) could be stored in a similar way to provide the possible of continuous community-driven quality control and to avoid repetition of data curation, which has to be done over and over again in a very subjective manner when the original data is used.
2) Describing Data Based on a Semantic Interoperability Layer
The first step described the recommendations on how to provide useful data and metadata. However, this still doesn’t solve the major challenge of data sharing following the FAIR approach, which is how to provide a mechanism to describe data and metadata in a structured format that is easy to understand by other users of the data or even by machines and, at the same time, provides flexibility and adaptability to new situations. In our opinion, these, on first sight, contradicting goals cannot be achieved by data/metadata standards alone. While more specific databases like ArrayExpress, TG-GATEs, and ChEMBL(concentrating on a very specific type of data) may require data in a very specific format, more general databases like PubChem (here we only refer to database part of PubChem and not the curated knowledge resources, a concept for the semantic annotation of PubChem compound and substance databases was already presented in Fu et al.) and BioStudies have to provide more open and flexible concepts. Both these resources were designed as data collection services linking out to the original data sources. For example, BioStudies is now hosting the diXa data warehouse, a study collections, where the study details are described in BioStudies with samples used and the actual technology specific data are being stored in Biosamples and in the various technology collections, respectively. However, nowadays BioStudies also offers storing capacities for data files not fitting in any of the specific databases without any requirements on the data format.
One approach we experiment with in OpenRiskNet is adding a semantic interoperability layer to existing databases. This is then used as a communication layer between users or modeling services and the databases. By using this interoperability layer, a database will be able to expose the following information:
- Scientific background of the dataset provided to the user to better understand the data and quality control, which can just be a link to the relevant publication but also to more detailed description including references to the studies and projects, in which they were generated, and to other related datasets (and for manuals, tutorials and other training material for software tools).
- So-called data and metadata schemata, i.e. the description of the content and the format of the stored data and the associated metadata, as well as options to access the data (search for data…) and terms of use.
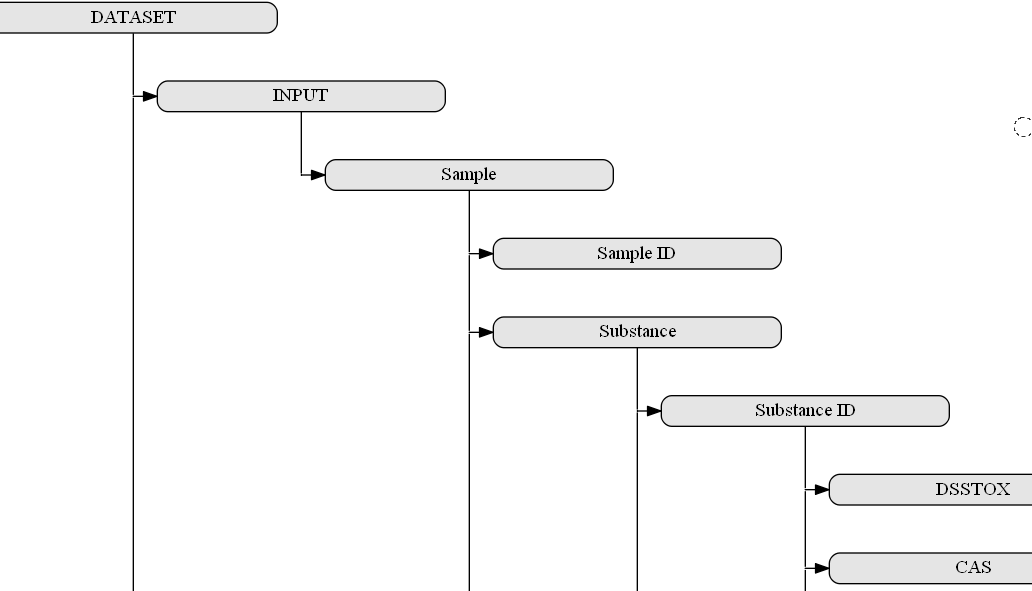
The most important aspect of the interoperability layer would be, on one hand, its flexibility with respect to the data types (and software tools), which can be covered, and, on the other hand, its consistency and clear rules with respect to the format the information is provided. The experimental setups are under constant development especially in respect to the recently introduced assays such as high content imaging/analysis and, therefore, the metadata and data schemata should allow for the integration of all important information as defined by the guidelines above. The proposed interoperability layer will give the data managers complete freedom over the structure of the provided data and associated functionalities but will also require them to describe it by using the interoperability layer ‘markup language’, which will increasingly rely on ontologies and reasoning based on these. We have started to create a concept for the semantic annotation of a dataset, which is grouping the metadata in the three sections “sample” (INPUT), “assay” (ACTION) and “results” (OUTPUT). It is somewhat derived from the ISA standard, which was designed to describe the process of investigation (I-tab, not included in our scheme because it is designed for a single dataset and not a complete investigation) leading to sample collections, on which specific studies (S-tab) are performed with assays (A-tab) that then yield results (not part of the ISA standard but separate files, which are referenced in the A-tab).
With the more general grouping in the brackets (INPUT->ACTION->OUTPUT), a similar semantic annotation approach can also be adopted for processing, analysis and modeling tools as a second group of services targeted for integration in the OpenRiskNet e-infrastructure. This is similar to the concept taken by bio.tools, which uses, beside others, the EDAM ontology for annotation, and in the API developed by the OpenRiskNet partner National Technical University of Athens for the Jaqpot Quattro modelling infrastructure. The INPUT of one of these in silico service would be the OUTPUT of data service or another computational tool and the required annotation and additional information, not directly needed by the downstream service, can directly be obtained by querying the upstream service. In the same way, the computational service should provide automatically the annotation needed for interpretation of the results and their usage in even additional modelling services or for scientific and regulatory reporting. In this way, the combination of all these semantic annotations of all used data services and tools will form a network similar to the RDF triple store, in total contributing to a full study (or investigation in the ISA namespace). A small part of such a network is shown in the figure below.
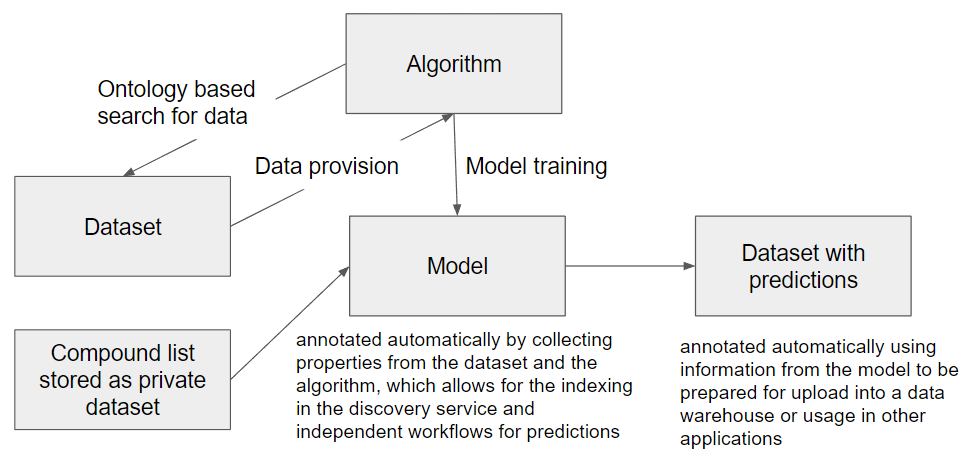
Even if the described concept builds a good starting point, many details have still to be worked out and technical solutions have to be developed. Fortunately, we are not the only ones working on the same goal and one of the next most important steps is to team up with 1) the ongoing work on the development of FAIR metrics, 2) the general data interoperability efforts done by the combined interoperability platform of ELIXIR, EXCELERATE and Corbel, and 3) the W3C Semantic Web for Health Care and Life Sciences (HCLS) Interest Group. If you like the approach and want to become part of the effort and contribute to a concept and technical realisation for data sharing, please stay tuned for updates in part 2 of this post or consider to become an associated partner of OpenRiskNet, giving you the chance to follow the developments at first hand and influence the direction we take.
About OpenRiskNet: openrisknet.org/about