The federation of computing and data resources between the OpenRiskNet e-infrastructure and public computational and data resources has now been demonstrated
The OpenRiskNet Consortium develops the OpenRiskNet e-infrastructure for the harmonisation and improved interoperability of data and software tools in the area of predictive toxicology and risk assessment. It aims to combine interoperable web services providing data or analysis, processing and modelling tools communicating over well-defined and harmonised application programming interfaces (APIs), supplemented by a semantic interoperability layer added to every service to describe the functionality whilst guaranteeing the technical and semantic interoperability.
An important task in the project, related also to the TGX case study, involved the federation of compute and data resources between the OpenRiskNet e-infrastructure and external resources. The reference environment has been designed to be capable of handling the majority of requirements for users wishes to deploy and run services. However specific situations demand solutions where either the computation, the data or both reside outside the OpenRiskNet e-infrastructure.
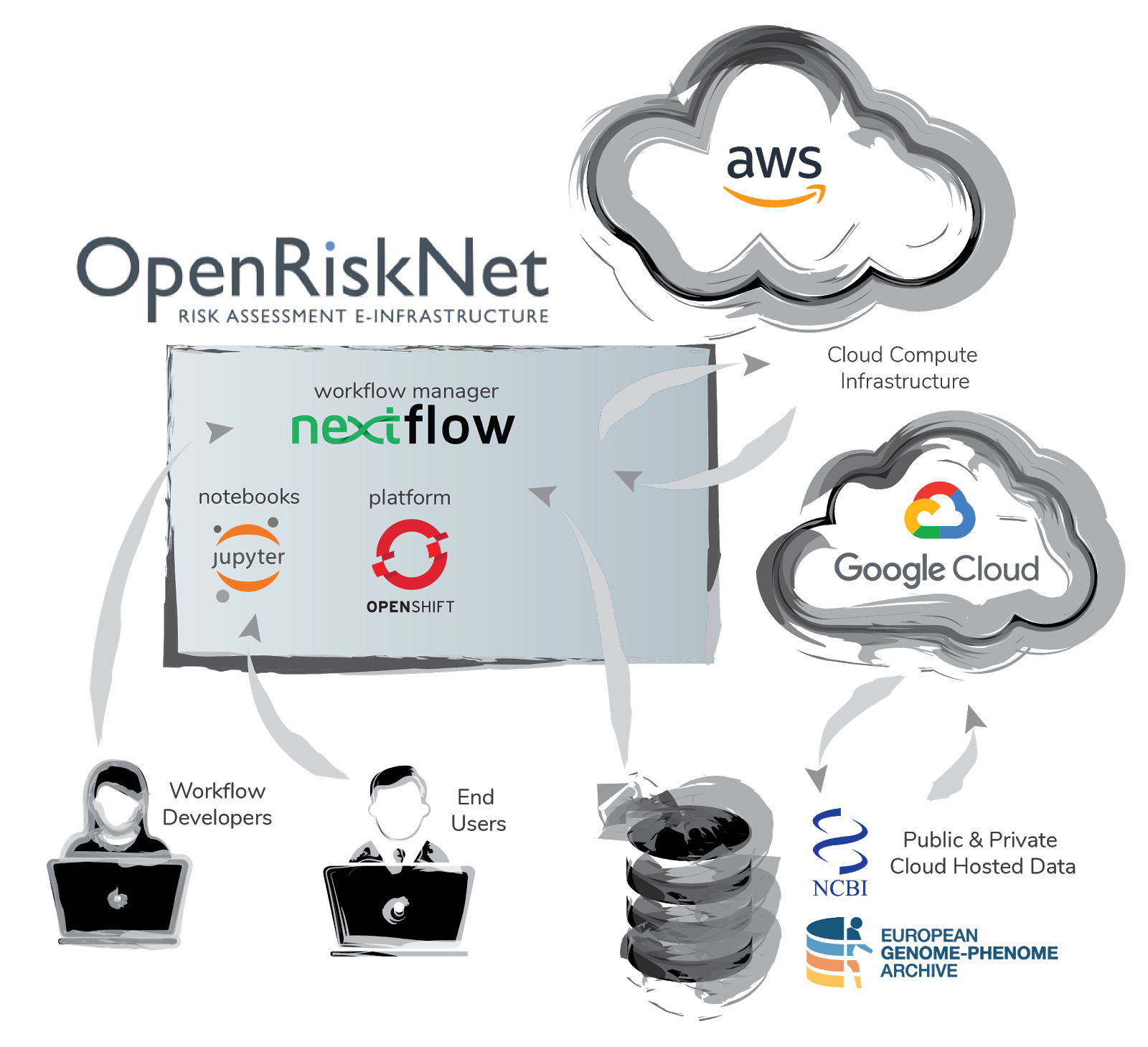
To illustrate the use of external resources we developed a toxicogenomics use case. The workflow termed “nf-toxomix” is a pipeline for toxicology predictions based on transcriptomic profiles. The workflow is built using Nextflow with an accompanying docker containers.
The workflow is adapted from Magkoufopoulou et al., 2012 research titled "A transcriptomics-based in vitro assay for predicting chemical genotoxicity in vivo". The method focuses on training the genotoxicity model with gene expression data from treated/non-treated samples and then assesses the prediction of genotoxicity using a training test validation approach. We expanded the workflow to include preprocessing steps where raw transcriptomic data is searched for and downloaded from the Sequence Read Archive, then mapped against the human reference genome and read counts generated. This step is computationally demanding and therefore was configured to be deployed on public cloud resources (AWS Batch) from the OpenRiskNet VRE.
The Nextflow Workflow Manager is a popular workflow manager for scientific data analysis workflows. Most importantly, Nextflow provides an abstraction layer between the application logic and the underlying execution platform that allows workflows to be completely portable. Scientists and developers write code in any scripting language, wrap the dependencies in containers or other popular package managers and have Nextflow orchestrate the tasks across different execution environments. A key difference of Nextflow is the dataflow programming model which uses a functional, data-driven and reactive approach which is capable of scaling large data applications consisting of millions of tasks.
To learn more on how this approach works, you are invited to join a live webinar demonstrating the use case and its technical implementation on 27 May 2019: Use of Nextflow tool for toxicogenomics-based prediction and mechanism identification in OpenRiskNet e-infrastructure.
Further readings:
Luechtefeld T, Marsh D, Rowlands C, Hartung T. Machine Learning of Toxicological Big Data Enables Read-Across Structure Activity Relationships (RASAR) Outperforming Animal Test Reproducibility. Toxicol Sci. 2018;165: 198–212. https://doi.org/10.1093/toxsci/kfy152
Di Tommaso P, Chatzou M, Floden EW, Barja PP, Palumbo E, Notredame C. Nextflow enables reproducible computational workflows. Nat Biotechnol. 2017;35: 316–319. https://doi.org/10.1038/nbt.3820
Köster J, Rahmann S. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics. Narnia; 2012;28: 2520–2522. https://doi.org/10.1093/bioinformatics/bts480
Magkoufopoulou C, Claessen SMH, Tsamou M, Jennen DGJ, Kleinjans JCS, van Delft JHM. A transcriptomics-based in vitro assay for predicting chemical genotoxicity in vivo. Carcinogenesis. 2012;33: 1421–1429. https://doi.org/10.1093/carcin/bgs182