Case Study
DataCure – Data curation and creation of pre-reasoned datasets and searching
Summary
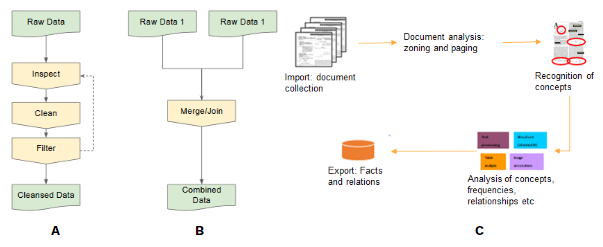
DataCure establishes a process for data curation and annotation that makes use of APIs (eliminating the need for manual file sharing) and semantic annotations for a more systematic and reproducible data curation workflow. In this case study, users are provided with capabilities to allow access to different OpenRiskNet data sources and target specific entries in an automated fashion for the purpose of identifying data and metadata associated with a chemical in general to identify possible areas of concern or for a specific endpoint of interest (Figure 1B). The datasets can be curated using OpenRiskNet workflows developed for this case study and, in this way, cleansed e.g. for their use in model development (Figure 1A). Text mining facilities and workflows are also included for the purposes of data searching, extraction and annotation (Figure 1C).
A first step in this process was to define APIs and provide the semantic annotation for selected databases (e.g. FDA datasets, ToxCast/Tox21 and ChEMBL). During the preparation for these use cases, it became clear that the existing ontologies do not cover all requirements of the semantic interoperability layer. Nevertheless, the design of the annotation process as an online or an offline/preprocessing step forms an ancillary part of this case study even though the ontology development and improvement cannot be fully covered by OpenRiskNet and is instead organized as a collaborative activity of the complete chemical and nano risk assessment community.
1. What we want to achieve?
Establish a process for data curation and annotation that makes use of APIs and semantic annotations for a more systematic and reproducible data curation workflow. The development of semantic annotations and API definition for selected databases are also desired.
2. What we want to deliver?
The aim is to demonstrate the access to OpenRiskNet data sources, deliver curated and annotated datasets for OpenRiskNet service users as well as preparation of and development of tools that can allow users to perform their own data curation.
3. What is the solution and how we implement?
Developing resources that can make use of APIs as much as possible and eliminate the need for manual file sharing. In addition, workflows that provide examples of useful data for predictive model generation, mechanistic investigations and toxicogenomic data analysis are developed.
Expected deliverables:
- Datasets accessible via APIs
- Data extraction and curation workflows
- Data annotation
Objectives
- This case study serves as the entry point of collecting and curating of all data sources to be used by the remaining use cases;
- The aim is to deliver curated and annotated datasets for OpenRiskNet service users as well as preparation and development of tools and workflows that allow users to perform their own data curation and analysis;
- Semantic annotation and API definition for the selected databases are also carried out in this use case.
DataCure covers the identification of chemical/substance of concern and collection of associated existing (meta)data. The steps in Tier 0 of the underlying risk assessment framework [ref] guide the data retrieval whenever applicable:
- Identification of molecular structure (if required);
- Collection of support data;
- Identification of analogues / suitability assessment and existing data;
- Identification/development of mapping identifiers;
- Collection of data for toxicology risk assessment.
Steps to achieve different objectives of the DataCure, include:
-
The user identifies and visualises the molecular structure:
- Generation of molecular identifiers for database search
- Searching all databases
- Data curation
- Tabular representation
- Visualisation
-
The user collects supporting data:
- Provide metadata including the semantically annotated dataschema using the interoperability layer providing information on the available data
- Access selected databases or flat files in a directory
- Query to ontology metadata service and select ontologies, which should be used for annotation
- Annotate and index all datasets using text mining extraction infrastructure
- Passing to ontology reasoning infrastructure
- Generate database of pre-reasoned dataset (semantic integration)
- Allow for manual curation
-
The user identifies chemical analogues:
- Inventory of molecules (commercially available or listed in databases)
- Generate list of chemically similar compounds
- Collect data of similar compounds
Databases and tools
The following set of data and tools are proposed to be used and exploited within the DataCure:
- Chemical, physchem, toxicological and omics databases: PubChem, registries (e.g. ECHA, INCI), EdelweissData Explorer (EwC), Liver Toxicology Knowledge Base (LTKB) and ChEMBL
- Cheminformatics tools to e.g. convert chemical identifiers, substructure and similarity searches: RDKit, CDK, Chemical Identifier Resolver (NIH), ChemidConvert and Fragnet Search
- Ontology/terminology/annotation: SCAIView API for semantic document retrieval, JProMiner/BELIEF UIMA components for concept tagging and normalisation, TeMOwl API for identifier/name to concept mapping
- Literature databases: PubMed, PubMed Central and ToxPlanet
Service integration
A set of physical-chemical properties, prediction, workflows, and ontology services are integrated including the SCAIView, TeMOwl, Jaqpot, Conformal prediction, and Jupyter notebooks.
Technical implementation
There are several steps involved in the technical implementation of this case study as listed below. All workflows mentioned here are made available to the public through the use of workflows written in scripting languages (such as R or Python) and prepared in Jupyter or Squonk notebooks. These workflows and notebooks are stored in the publicly accessible OpenRiskNet GitHub repository.
- Data sources: EdelweissData (further described below) serves as one of the main data provisioning tools for this case study and others in the project. There are also other data sources that are directly used such as ChEMBL, PubMed and ToxPlanet, where the latter two are repositories primarily storing literature and literature based data.
- Data Extraction: The retrieval of data from EdelweissData and all other resources are done through the use of API calls. Workflows with examples of various forms of data extraction using these APIs are documented and run from Jupyter and Squonk notebooks. Datasets can be retrieved for the query chemical and/or for similar compounds identified using the Fragnet search REST API. Additionally, these extractions partly also involve text mining using the SCAIView tool also integrated into the Jupyter notebooks.
- Data Searching: Workflows that employ text mining capabilities are used for searching for specific data and refinement and curation of the data extracted from these sources.
- Data curation and reasoning: This is done through the provision of workflows stored in Jupyter or Squonk notebooks. These workflows cover components such as extraction of specific data and merging of datasets for downstream analysis purposes.
- Resubmission to data source: Even if not implemented during the OpenRiskNet project, curated datasets may be resubmitted to an OpenRiskNet-compliant data management solution like EdelweissData to be used by others through API access.
Description of tools mentioned in technical implementation
- EdelweissData: This platform is a web based data management tool, which is used to provide multiple OpenRiskNet data sources like ToxCast/Tox21, TG-GATEs and the Daphnia dataset. It gives users the ability to filter, search and access data based on rich metadata through an API.
- Jupyter Notebooks: This is an open-source web application that provides an interactive programming environment to create, share, and collaborate on code and other documents.
- Squonk Computational Notebooks: This is an open-source web application provided by IM that is somewhat similar in concept to Jupyter, but targeted at the scientist rather than the programmer.
- SCAIView: This is an text-mining and information retrieval tool that uses semantic and ontological searches to extract relevant information from a variety of unstructured textual data sources. It can work online or offline to access both publicly available or proprietary data sources available in various formats. In the academic version three large datasets are pre-annotated: Medline (~29 mio documents), PMC (2.6 mio) and US patent corpus (4.4 mio).
- TeMOwl: This service provides unified access to semantic data i.e. controlled vocabularies, terminologies, ontologies and knowledge resources. It hides complexity of different semantic service providers including their various data formats. Further it aggregates (integrates, maps or aligns) different information resources. Concepts, things of thought, are often defined within multiple resources, even though they refer to the same thing. The system tries to unify those. TeMOwl is an API Service, that offers programmatic access to semantic information.
- Fragnet Search: This is a chemical similarity search REST API that uses the Fragment Network (conceived by Astex Pharmaceuticals in this paper) to identify related molecules. The typical use of the API is to specify a query molecule and get back a set of molecules that are related to it according to some search parameters, and so is a good approach for “expanding” out one or more molecules to get other related molecules to consider. The Fragment Network is more effective and chemically meaningful compared to traditional fingerprint based similarity search techniques especially for small molecules. More information can be found here.
Outcomes
Outcome from this case study provide example workflows to illustrate the processes described above and the extracted datasets with accompanying metadata were used in the AOPLink, TGX and ModelRX case studies and testing of the services integrated therein. They can now be easily adapted to real problem scenario supporting data collection in chemical risk assessment.
Details on the outcomes, are included in the case study report linked below and refer to:
- Data access and curation workflow using Squonk
- Data curation for the LTKB dataset
- Merging LTKB data with TG-GATEs
- Combining of data and knowledge sources
- Finding similar data-rich compounds for read across
- Data mining workflow for carcinogenicity predictions
- DataCure webinar
Currently available services:
-
Service type: Database / data source
-
ToxCast and Tox21 datasets (raw and summary) extracted from the MySQL database provided by US EPAService type: Database / data source
-
Our platform searches the content from 500+ websites and quickly delivers relevant chemical hazard and toxicology data.Service type: Database / data source, Data mining tool, Service
-
Collection of toxicological data sources exposed via OpenToxService type: Database / data source, Application, Service
-
Python client for Squonk REST APIService type: Software
-
Extend molecular biological networks with toxic compounds.Service type: Database / data source
-
Service type: Database / data source
-
TeMOwl provides unified access to semantic data i.e. controlled vocabularies, terminologies, ontologies and knowledge resources.Service type: Helper tool, Software, Service
-
Chemical similarity using the Fragment NetworkService type: Database / data source, Service
-
Processed data (counts and fold changes) based on the transcriptomics data provided by Open TG-GATEs and DrugMatrixService type: Database / data source
-
Chemical identifier conversion serviceService type: Helper tool
-
Information retrieval system for semantic searches in large text collectionsService type: Application, Software, Service
-
Interactive computing and workflows sharingService type: Visualisation tool, Helper tool, Software, Analysis tool, Processing tool, Workflow tool
-
Computation research made simple and reproducibleService type: Database / data source, Visualisation tool, Software, Analysis tool, Service, Workflow tool
Related resources
The OpenRiskNet case studies (originally outlined in Deliverable 1.3) were developed to demonstrate the modularised application of interoperable and interlinked workflows. These workflows were designed to address specific aspects required to inform on the potential of a compound to be toxic to humans and to eventually perform a risk assessment analysis. While each case study targets a specific area including data collection, kinetics modelling, omics data and Quantitative Structure Activity Relationships (QSAR), together they address a more complete risk assessment framework. Additionally, the modules here are fine-tuned for the utilisation and application of new approach methodologies (NAMs) in order to accelerate the replacement of animals in risk assessment scenarios. These case studies guided the selection of data sources and tools for integration and acted as examples to demonstrate the OpenRiskNet achievements to improve the level of the corresponding APIs with respect to harmonisation of the API endpoints, service description and semantic annotation.
DataCure establishes a process for data curation and annotation that makes use of APIs (eliminating the need for manual file sharing) and semantic annotations for a more systematic and reproducible data curation workflow. In this case study, users are provided with capabilities to allow access to different OpenRiskNet data sources and target specific entries in an automated fashion for the purpose of identifying data and metadata associated with a chemical in general to identify possible areas of concern or for a specific endpoint of interest (Figure 1B). The datasets can be curated using OpenRiskNet workflows developed for this case study and, in this way, cleansed e.g. for their use in model development (Figure 1A). Text mining facilities and workflows are also included for the purposes of data searching, extraction and annotation (Figure 1C). A first step in this process was to define APIs and provide the semantic annotation for selected databases (e.g. FDA datasets, ToxCast/Tox21 and ChEMBL). During the preparation for these use cases, it became clear that the existing ontologies do not cover all requirements of the semantic interoperability layer. Nevertheless, the design of the annotation process as an online or an offline/preprocessing step forms an ancillary part of this case study even though the ontology development and improvement cannot be fully covered by OpenRiskNet and is instead organized as a collaborative activity of the complete chemical and nano risk assessment community.
Report
The OpenRiskNet project (https://openrisknet.org/) is funded by the H2020-EINFRA-22-2016 Programme. Here we present how the concept of Adverse Outcome Pathways (AOPs), which captures mechanistic knowledge from a chemical exposure causing a Molecular Initiating Event (MIE), through Key Events (KEs) towards an Adverse Outcome (AO), can be extended with additional knowledge by using tools and data available through the OpenRiskNet e-Infrastructure. This poster describes how the case study of AOPLink, together with DataCure, TGX, and SysGroup, can utilize the AOP framework for knowledge and data integration to support risk assessments. AOPLink involves the integration of knowledge captured in AOPs with additional data sources and experimental data from DataCure. TGX feeds this integration with prediction models of the MIE of such AOPs using either gene expression data or knowledge about stress response pathways. This is complemented by SysGroup, which is about the grouping of chemical compounds based on structural similarity and mode of action based on omics data. Therefore, the combination of these case studies extends the AOP knowledge and allows biological pathway analysis in the context of AOPs, by combining experimental data and the molecular knowledge that is captured in KEs of AOPs.
ToxTargetLinks
EdelweissData serving ToxCast, ToxRefDB and TG-GATEs data
Slides
Example workflow based on OpenRiskNet tools - Pathway identification workflow related to DataCure and AOPlink case studies. This notebook downloads TG-Gates data of 4 compounds and selects genes overexpressed in all sample. The Affymetrix probe sets are then translated into Ensembl gene identifiers using the BridgeDB service and pathways associated with the genes are identified using the WikiPathways service.
BridgeDb identifier mapping service
EdelweissData serving ToxCast, ToxRefDB and TG-GATEs data
Jupyter Notebooks