Case Study
MetaP – Metabolism Prediction
Summary
Metabolites may well play an important role in adverse effects of parent drug or other xenobiotic compounds. In this case study VU (CS leader), HITeC/HHU (associate partner and implementation challenge winner), JGU, and UU have worked together on making methods and tools available for metabolite and site-of-metabolism (SOM) prediction. For that purpose we integrated and used ligand-based metabolism predictors (e.g. MetPred, enviPath, FAME, SMARTCyp) and we incorporated protein-structure and -dynamics based approaches to predict SOMs by Cytochrome P450 enzymes (P450s). P450s metabolise ~75% of the currently marketed drugs and their active-site shape and plasticity often play an important role in determining the substrate’s SOM. It is expected that this work will be continued after the end of the project to make services available for the prediction of microbial biotransformation pathways by integrating the enviPath data and software developed in part by JGU.
During method development, model calibration and validation we used databases such as XMetDB and other open-access databases for drugs, xenobiotics and their respective metabolites. To facilitate the combined use of the metabolite prediction approaches and their outcomes, we benefited of ongoing development in workflow management systems and we made Jupyter Notebooks available to facilitate collection and visualization of results from the different available services. We illustrated the added value of having multiple predictors and our Jupyter notebooks available, in a pilot study on retrospective consensus predictions of known SOMs for drug compounds for which possible metabolite-associated toxicity was previously reported.
Objectives
The objective of this case study was to enable and facilitate metabolite prediction within the OpenRiskNet infrastructure and to evaluate and demonstrate the added value of it. For that purpose we integrated different tools for metabolism prediction, including tools for:
- Ligand-based site-of-metabolism (SOM) prediction using reaction SMARTs, circular fingerprints and/or atomic reactivities;
- QSBR (quantitative-structure biotransformation relationship) modeling of microbial biotransformation;
- Protein-structure and -dynamics based prediction of CYP450 isoform specific binding and SOMs;
- Predicting probabilities for specific reaction type events.
Combined use of the tools has been made possible and compared using Jupyter notebooks that gather and visualize results from the available case-study services.
See the “Databases and tools” subsection for more details on the corresponding tools. For our comparisons of predictive (and consensus) performance we used selected compounds from literature for which SOMs and metabolite-associated toxicity have been reported. We anticipate to present our results in an upcoming manuscript on tool integration, which will illustrate how using several tools can have additional value (when compared to individual tools) to (site-of-)metabolism prediction.
Risk assessment framework
Prediction outcomes can serve as input for other molecular structure-based AO predictors, which relates to Tier 0 (Step 1: identification of molecular structure) and Tier 1 (Step 6: mechanism of action).
Databases and tools
The table below gives an overview of metabolite prediction tools that are integrated and have been used in this case study. During method development, model calibration, and validation, advantage was taken of data from XMetDB (ref.) and other databases for drugs, xenobiotics and their respective metabolites, as available in ZINC, ChEMBL, DrugBank, EAWAG-BBD and/or the SMARTCyp and FAME suites. Integration of enviPath is still ongoing, which is a database and prediction system for microbial biotransformation of organic environmental contaminants.
| Tool | Input | Ouput | Method | MetPred | 2D chemical structure of ligand | SOMs with Reaction Types for Phase I reactions | Preprocess Metabolite reaction database (>100K biotransformations) using MCS. For each query compound, look up similar atom environments based on circular fingerprints and use ReactionSMARTS to identify reaction types. See (ref). |
| FAME 3 | 2D chemical structure of ligand | SOMs for Phase I, Phase II, or combined Phase I/II metabolism | Machine learning using 2D-circular-environment based atomic descriptors. See (ref). |
| SMARTcyp 2.0 | 2D chemical structure of ligand | Rank atoms (SOMs) for P450-isoform specific reactions | Combining reactivity (from database on QM calculated transition state energies) with simple 2D molecular accessibility descriptors for SOM prediction. See (ref). |
| Plasticity tools | 3D Chemical structure of ligand | Prediction of most probable SOMs for P450-isoform specific reactions | Protein-structure and dynamics based prediction of substrate binding orientations and corresponding SOM in the active site of CYP isoforms (1A2, 2D6, 3A4). See (ref). |
Technical implementation
As summarised in the table above, several services have come available in the MetaP case study. The listed services offer their functionality through RESTful APIs that are formalised according to OpenAPI specifications. The APIs are build using the Swagger toolchain and subsequently enable direct user interaction with the API endpoints using a browser-based User Interface (the Swagger UI). In addition, MetPred and SMARTCyp offer a custom browser-based interface to their service. The APIs enable access to the core features of the services as summarised above, and typically accept submissions of chemical structures in common file formats.
API endpoint input and output data exchange is standardised to a machine-readable JSON format. Together with the OpenAPI data type definitions and JSON-LD data annotation it ensures seamless integration of the containerised services in the OpenRiskNet infrastructure and data exchange with other services.
Service API use and interoperability of the listed services is demonstrated using a Jupyter Notebook freely available in GitHub. Single 3D ligand structures in Tripos MOL2 format are used as input to the various services and the standardised JSON output are aggregated into a Pandas DataFrame demonstrating interoperability. Predicted SOMs are visualized on the 2D ligand depiction using the RDKit package.
Outcomes
In addition to the service integration of the metabolite prediction tools listed above, we have evaluated the added value of having multiple tools and their combined use available (via Jupyter Notebooks). The different predictors give complementary types of output. MetPred, FAME 3, and SMARTCyp tools predict SOMs related to Phase I, Phase I/II, and Cytochrome P450 isoform specific conversion, respectively. Per (heavy) atom, normalized propensities are written out to indicate the likelihood of the atom to be a SOM. In addition, MetPred also gives back most probable reaction types at predicted SOMs. Facilitated by the Jupyter Notebook that supplies and visualizes output from the different predictors (see the case study report linked below), the MetaP tools can thus aid experts in guiding decision making on metabolite formation and/or in obtaining input for subsequent case studies.
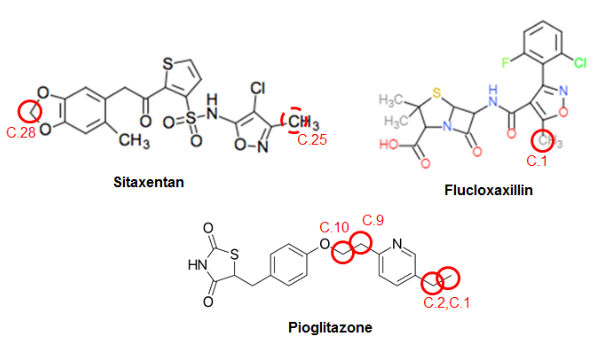
The added value of having the multiple complementary tools available for metabolite prediction is illustrated by the Jupyter-notebook output presented in the case study report, which collects SOM predictions and MetPred predictions of Phase I reaction types (and which color-highlights atoms as predicted SOM if propensities are larger than a preset cutoff) for the three compounds (see the Figure below). These compounds were selected because possible toxicological effects have been related with their metabolites, and their metabolism is extensively studied in literature.
Currently available services:
-
Python client for Squonk REST APIService type: Software
-
Chemical similarity using the Fragment NetworkService type: Database / data source, Service
-
Predict ADME/PK with ConfidenceService type: Application, Software, Service
-
Machine learning models for site-of-metabolism predictionService type: Application, Software, Trained model, Model, Service
-
Webservice to WEKA Machine Learning AlgorithmsService type: Trained model, Model generation tool, Model, Service
-
Interactive computing and workflows sharingService type: Visualisation tool, Helper tool, Software, Analysis tool, Processing tool, Workflow tool
-
Service type: Trained model, Service
-
Service type: Application, Software, Processing tool, Trained model, Service
-
Computation research made simple and reproducibleService type: Database / data source, Visualisation tool, Software, Analysis tool, Service, Workflow tool
Related resources
The OpenRiskNet case studies (originally outlined in Deliverable 1.3) were developed to demonstrate the modularised application of interoperable and interlinked workflows. These workflows were designed to address specific aspects required to inform on the potential of a compound to be toxic to humans and to eventually perform a risk assessment analysis. While each case study targets a specific area including data collection, kinetics modelling, omics data and Quantitative Structure Activity Relationships (QSAR), together they address a more complete risk assessment framework. Additionally, the modules here are fine-tuned for the utilisation and application of new approach methodologies (NAMs) in order to accelerate the replacement of animals in risk assessment scenarios. These case studies guided the selection of data sources and tools for integration and acted as examples to demonstrate the OpenRiskNet achievements to improve the level of the corresponding APIs with respect to harmonisation of the API endpoints, service description and semantic annotation.
Metabolites may well play an important role in adverse effects of parent drug or other xenobiotic compounds. In this case study VU (CS leader), HITeC/HHU (associate partner and implementation challenge winner), JGU, and UU have worked together on making methods and tools available for metabolite and site-of-metabolism (SOM) prediction. For that purpose we integrated and used ligand-based metabolism predictors (e.g. MetPred, enviPath, FAME, SMARTCyp) and we incorporated protein-structure and -dynamics based approaches to predict SOMs by Cytochrome P450 enzymes (P450s). P450s metabolise ~75% of the currently marketed drugs and their active-site shape and plasticity often play an important role in determining the substrate’s SOM. It is expected that this work will be continued after the end of the project to make services available for the prediction of microbial biotransformation pathways by integrating the enviPath data and software developed in part by JGU. During method development, model calibration and validation we used databases such as XMetDB and other open-access databases for drugs, xenobiotics and their respective metabolites. To facilitate the combined use of the metabolite prediction approaches and their outcomes, we benefited of ongoing development in workflow management systems and we made Jupyter Notebooks available to facilitate collection and visualization of results from the different available services. We illustrated the added value of having multiple predictors and our Jupyter notebooks available, in a pilot study on retrospective consensus predictions of known SOMs for drug compounds for which possible metabolite-associated toxicity was previously reported.
Report
JGU WEKA REST Service
Metabolites can play an important role in adverse effects of parent drug (or other xenobiotic) compounds. During the EU-H2020 OpenRiskNet project, several partners (VU Amsterdam, HHU/HITeC Hamburg, Uppsala University, JGU Mainz) have worked together on making methods and tools available within the OpenRiskNet platform for metabolite and site-of-metabolism (SOM) prediction. For that purpose we have integrated ligand-based metabolite predictors (e.g., MetPred, FAME 3, SMARTCyp) and protein-structure and -dynamics based models to predict SOMs of Cytochrome P450 (CYP450) substrates. CYP450s metabolize ~75% of the currently marketed drugs and their active-site shape and plasticity often play an important role in determining the substrate's SOM. To facilitate the combined use of the metabolite prediction approaches and their outcomes, we made Jupyter notebooks available that gather and visualize results from the integrated services. Here we illustrate the possible added value of their combined use in the context of a pilot study on SOM prediction for compounds with known metabolite-associated toxicity. Finally we shortly discuss related work from our laboratory, on predicting Cytochrome P450 binding affinity prediction.