Case Study
ModelRX – Modelling for Prediction or Read Across
Summary
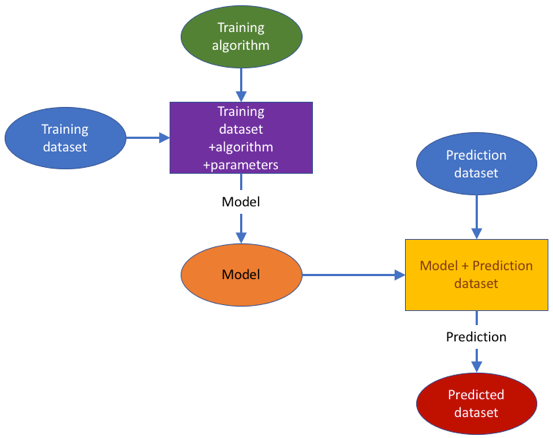
The ModelRX case study was designed to cover the important area of generating and applying predictive models, and more specifically QSAR models in hazard assessment endorsed by different regulations, as completely in silico alternatives to animal testing and useful also in early research when no data is available for a compound. The QSAR development process schematically presented in the figure below, begins by obtaining a training data set from an OpenRiskNet data source. A model can then be trained with OpenRiskNet modelling tools and the resulting models are packaged into a container, documented and ontologically annotated. To assure the quality of the models, they are validated using OECD guidelines (Jennings et al. 2018). Prediction for new compounds can be obtained using a specific model or a consensus of predictions of all models. This case study will present this workflow with the example of blood-brain-barrier (BBB) penetration, for which multiple models were generated using tools from OpenRiskNet consortium and associated partners used individually as well as in a consensus approach using Dempster-Shafer theory (Park et al. 2014; Rathman et al. 2018).
Objectives
The objectives of this case study are: use in silico predictive modelling approaches (QSAR) to support final risk assessment by supporting similarity identification related to the DataCure case study (by providing tools for calculating theoretical descriptors of substances) and fill data gaps for specific compounds or to augment incomplete datasets.
Risk assessment framework
The ModelRX case study contributes in two tiers (as tiers are defined in Berggren et al. 2017:
- On the one hand, it provides computational methods to support suitability assessment of existing data and identification of analogues (Tier 0);
- On the other hand, it provides predictive modelling functionalities, which are essential in the field of final risk assessment (Tier 2).
Databases and tools
Jaqpot Quattro (NTUA), CPSign (UU), JGU WEKA Rest service (JGU), Lazar (JGU/IST).
Technical implementation
From the developer’s perspective, this case study demonstrates the improved interoperability and compatibility/complementarity of the models based on services deployed following the general steps that have been agreed for developing the OpenRiskNet infrastructure and services. Each application is delivered in the form of a container image and deployed. Docker is used as the basic engine for the containerisation of the applications. Above that, OpenShift, which is a container orchestration tool, is used for the management of the containers and services. OpenShift provides many different options for deploying applications. Some recipes and examples have been documented in the OpenRiskNet GitHub page.
When an application is deployed, a service discovery mechanism is responsible for discovering the most suitable services for each application. Based upon the OpenAPI specification, each API should be deployed with the swagger definition. This swagger file should then be integrated with the Json-LD annotations as dictated by the Json-LD specification. The discovery service mechanism parses the resulting Json-LD and resolves the annotations into RDF triplets. These triplets can then be queried with SPARQL. The result of the SPARQL query lets the user know which services are responsible for making models or predictions. The documentation can be found via swagger definition of each application. This way, the services are integrated into the OpenRiskNet virtual environments and can be used and incorporated into end user applications and other services as demonstrated here with a workflow performing consensus modelling.
QSAR modelling was already the main topic of the OpenTox project, which is a clear predecessor of OpenRiskNet. OpenTox had a much more focused aim and the clear goal to have very interlinked services, where it is even possible to combine parts of the workflow from different partners, e.g. descriptor calculation is performed by a service from one partner, the model is trained using algorithms from another partner, and finally the prediction is performed by integrating the trained model into a user interface of a third partner. To allow this, the technical implementation had to be based on rigorously defined modelling APIs the OpenTox standards. Additionally, following these specifications and standards opened the full flexibility of model development and performing predictions to the user, which, on the one side, allows optimization of the workflow to a specific problem at hand but, on the other hand, also requires more experienced users. Therefore, the case study and the integration of QSAR services into OpenRiskNet in general used the OpenTox specifications as the starting point for developing a more flexible QSAR workflow, which, like the OpenTox workflow, has all components represented in the figure above, but allows services to combine and simplify different steps and, in this way, e.g. reduces the necessity for rigorous usage of Uniform Resource Identifiers (URIs) for simple substances, which can be referenced by chemical identifiers like SMILES or InChIs, and descriptors, e.g. when they are calculated on the fly by the service. This allows the easier integration of external tools as e.g. provided by the associated partners, which don’t support all the features required for an OpenTox service. This approach allows us to include both more straightforward tools that can work together with OpenRiskNet with minimal setup, but at the same time can accommodate more feature-rich tools that require more feature-rich APIs to provide their full offering.
However, OpenRiskNet enforces additional requirements due to the broadening of the application area. As described in detail in deliverable reports D2.2 and D2.4, standardization of the APIs was not possible and even not desired to allow very different tools from many areas to be run on the OpenRiskNet virtual environments. Instead, harmonization and interoperability was obtained by semantic annotation of the APIs and the semantic interoperability later, which provide the user with the information needed to link to services using workflow tools. Modelling APIs need a high level of integration into the OpenRiskNet ecosystem. Integration with the DataCure CS is vital. On the semantic interoperability layer, training datasets should be compatible with an algorithm and, in turn, prediction datasets should be compatible with a prediction model. Additionally, in a best practice scenario, the generated models and datasets need to be accompanied with semantic metadata on their life cycle, thus enforcing semantic enrichment of the dynamically-created entities. Algorithms, models and predicted datasets are built as services, discoverable by the OpenRiskNet discovery service. This is a step that should occur whenever an entity (algorithm, model, predicted dataset) is created.
To enable the user to train models and use them in predictions, , the guidelines agreed on by OpenRiskNet for functionality, which should be provided by QSAR and read-across services to allow highest flexibility include the following steps. As already stated before, some of the requirements might become irrelevant for a specific service if it combines different steps to provide an easier way to make predictions, especially for less experienced users. More details on the features implemented in each service are annexed below.
- Selecting a training data set
The user chooses among OpenRiskNet compliant data sets already accessible through the discovery service. Following the OpenTox specification, a Dataset includes, at a minimum:
- a dataset URI
- substances: substance URIs (each substance URI will be associated with a term from the ontology)
- features:
- feature URIs (each feature URI will be associated with a term from the ontology)
- values in numerical format
- category (experimental/computed)
- if computed, the URI of the model used to generate the values
- Units
An alternative path to provide data in OpenRiskNet e.g. followed in the DataCure case study is by dedicated services offering the following elements:
- a dataset URI
- a semantically annotated data API providing information on how to access the data and what specific data schema is used
With this information, it will be possible for alternative implementations to integrate into OpenRiskNet and interact with modelling services following the full OpenTox specifications, provided they also implement intermediate processing steps, i.e. within the environment of a Jupyter notebook, to structure the data so that it fulfills the minimum set of requirements for the following steps of the QSAR workflow.
- Selecting a (suitable) modelling algorithm
The user will be able to choose from a list of suitable algorithms. Algorithms should include at a minimum:
- algorithm URI
- title
- description
- algorithm type (regression/classification)
- default values for its parameters (where applicable)
- Specifying parameters and Generating Predictive model
Once an algorithm has been selected, the user defines the endpoint, selects the tuning parameters, (only if different values from the default ones are desired) and runs the algorithm. The generated Model contains, at a minimum:
- model URI
- title
- description
- the URI of the dataset that was used to create it
- the URIs of the input features
- the URI of the predicted feature
- values of tuning parameters
The following were identified as possible extensions:
- Include services/APIs for validation of the generated model
- Provide mechanisms to pick out the best algorithm for a specific dataset: (e.g. RRegrs)
- Include algorithms to calculate domain of applicability
- Selecting a prediction data set
After the creation of a model, the user selects a prediction dataset meeting all the requirements specified in (Chomenidis et al. 2017). This dataset is tested for compatibility against the required features of the model in terms of feature URIs, i.e. the dataset should contain all the subset of features used to produce the model. Additional features are allowed, however they will be ignored.
- Running predictive model on the prediction data set
The predictive model is applied on the prediction dataset to generate the predicted dataset, which must be compatible with the requirements specified in (Chomenidis et al. 2017). The predicted dataset augments the prediction dataset with all necessary information about the predicted feature:
- prediction feature URIs (each feature URI will be associated with a term from the ontology)
- values in numerical format
- category (computed)
- the URI of the model used to generate the values
- Units
Examples of implementation are linked below under related resources.
Outcomes
The work on the case study was designed to showcase how the workflow defined above for producing semantically annotated predictive models can be shared, tested, validated and eventually be applied for predicting adverse effects of substances in a safe by design and/or risk assessment regulatory framework. OpenRiskNet provides the necessary functionalities that allow not only service developers but also researchers and practitioners to easily produce and expose their models as ready-to-use web applications. The OpenRiskNet e-infrastructure serves as a central model repository in the area of predictive toxicology. For example, when a research group publishes a predictive model in a scientific journal, they can additionally provide the implementation of the model as a web service using the OpenRiskNet implementation. The produced models contain all the necessary metadata and ontological information to make them easily searchable by the users and systematically and rigorously define their domain of applicability. Most importantly, the produced resources are not just static representations of the models, but actual web applications where the users can supply the necessary information for query substances and receive the predictions for their adverse effects. However, because of the harmonization and interoperability, these are not just stand-alone tools but can be easily combined to improve the overall performance or can be used to replace older tools with newer ones without changing the overall procedure. This was demonstrated with the workflows for developing a consensus model for blood-brain-barrier (BBB) penetration (available online). The test set of 414 compounds was obtained from the (Lazar service).
Details on the outcomes are included in the case study report linked below.
Currently available services:
-
Service type: Trained model
-
Korea Institute of Technology NanoQSAR modelService type: Trained model, Model
-
The On-line Chemical Database and Modeling Environment modelsService type: Trained model
-
Virtual Screening Metal Oxide Nanoparticles Enalos PlatformService type: Application, Trained model, Model, Service
-
Enalos Platform nanoinformatics nanoparticlesService type: Trained model, Model
-
Generate, store and share predictive statistical and machine learning modelsService type: Analysis tool, Processing tool, Trained model, Model generation tool, Model, Data mining tool, Service
-
Generate, store and share predictive statistical and machine learning modelsService type: Application, Visualisation tool, Analysis tool, Processing tool, Trained model, Model generation tool, Model, Data mining tool, Workflow tool
-
Toxicity predictionsService type: Application, Helper tool, Trained model, Model, Service
-
Python client for Squonk REST APIService type: Software
-
Service type: Database / data source
-
Chemical similarity using the Fragment NetworkService type: Database / data source, Service
-
A classification model built by CPSign for predicting moleculesService type: Trained model, Model
-
A Venn-ABERS model built by CPSign for predicting moleculesService type: Trained model, Model
-
Service type: Trained model
-
Service type: Trained model
-
Webservice to WEKA Machine Learning AlgorithmsService type: Trained model, Model generation tool, Model, Service
-
Interactive computing and workflows sharingService type: Visualisation tool, Helper tool, Software, Analysis tool, Processing tool, Workflow tool
-
Computation research made simple and reproducibleService type: Database / data source, Visualisation tool, Software, Analysis tool, Service, Workflow tool
Related resources
The OpenRiskNet case studies (originally outlined in Deliverable 1.3) were developed to demonstrate the modularised application of interoperable and interlinked workflows. These workflows were designed to address specific aspects required to inform on the potential of a compound to be toxic to humans and to eventually perform a risk assessment analysis. While each case study targets a specific area including data collection, kinetics modelling, omics data and Quantitative Structure Activity Relationships (QSAR), together they address a more complete risk assessment framework. Additionally, the modules here are fine-tuned for the utilisation and application of new approach methodologies (NAMs) in order to accelerate the replacement of animals in risk assessment scenarios. These case studies guided the selection of data sources and tools for integration and acted as examples to demonstrate the OpenRiskNet achievements to improve the level of the corresponding APIs with respect to harmonisation of the API endpoints, service description and semantic annotation.
The ModelRX case study was designed to cover the important area of generating and applying predictive models, and more specifically QSAR models in hazard assessment endorsed by different regulations, as completely in silico alternatives to animal testing and useful also in early research when no data is available for a compound. The QSAR development process schematically presented in Figure 1 begins by obtaining a training data set from an OpenRiskNet data source. A model can then be trained with OpenRiskNet modelling tools and the resulting models are packaged into a container, documented and ontologically annotated. To assure the quality of the models, they are validated using OECD guidelines (Jennings et al. 2018). Prediction for new compounds can be obtained using a specific model or a consensus of predictions of all models. This case study will present this workflow with the example of blood-brain-barrier (BBB) penetration, for which multiple models were generated using tools from OpenRiskNet consortium and associated partners used individually as well as in a consensus approach using Dempster-Shafer theory (Park et al. 2014; Rathman et al. 2018).
Report
Nano-QSAR to predict cytotoxicity of metal and metal oxide nanoparticles
Lazar Toxicity Predictions
JGU WEKA REST Service
The OpenRiskNet project (https://openrisknet.org/) is funded by the H2020-EINFRA-22-2016 Programme and its main objective is the development of an open e-infrastructure providing data and software resources and services to a variety of industries requiring risk assessment (e.g. chemicals, cosmetic ingredients, pharma or nanotechnologies). The concept of case studies was followed in order to test and evaluate proposed solutions and is described in https://openrisknet.org/e-infrastructure/development/case-studies/. Two case studies, namely ModelRX and RevK, focus on modelling within risk assessment. The ModelRX – Modelling for Prediction or Read Across case study provides computational methods for predictive modelling and support of existing data suitability assessment. It supports final risk assessment by providing calculations of theoretical descriptors, gap filling of incomplete datasets. computational modelling (QSAR) and predictions of adverse effects. Services are offered through Jaqpot (UI/API), JGU WEKA (API), Lazar (UI) and Jupyter & Squonk Notebooks. In the RevK – Reverse dosimetry and PBPK prediction case study, physiologically based pharmacokinetic (PBPK) models are made accessible for the purpose of risk assessment-relevant scenarios. The PKSim software, the httk R package and custom-made PBPK models have been integrated. RevK offers services through Jaqpot (UI/API).
Nano-QSAR to predict cytotoxicity of metal and metal oxide nanoparticles
Lazar Toxicity Predictions
Lazar Toxicity Predictions
Slides
Lazar Toxicity Predictions
Nano-QSAR to predict cytotoxicity of metal and metal oxide nanoparticles
Jaqpot API
Jaqpot GUI