Case Study
SysGroup – A systems biology approach for grouping compounds
Summary
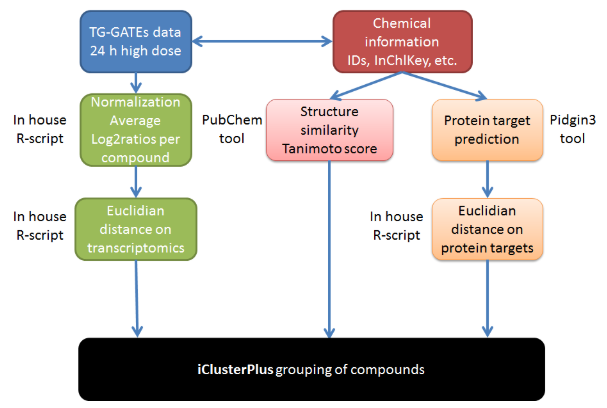
This case study used the approach of the diXa / DECO2 (Cefic-LRI AIMT4) projects to reproduce and extend the results obtained on the identification of hepatotoxicant groups based on similarity in mechanisms of action (omics-based) and chemical structure using services from OpenRiskNet. The figure below displays the workflow that was used in this case study: workflow for grouping compounds by integrating transcriptomics data, Tanimoto similarity scores and ligand-based target predictions using iClusterPlus.
Objectives
The objective of this case study is to implement an integrated analysis using chemoinformatics and omics data for improved grouping of compounds with similar toxicity and/or mode of action.
Risk assessment framework
SysGroup covers the identification of use scenario / chemical of concern / collection of existing information (Tier 0 in the selected framework) and its steps related to:
- Identification of molecular structure;
- Collection of support data;
- Identification of analogues / suitability assessment and existing data.
Databases and tools
In this case study TG-GATEs transcriptomics data from primary human hepatocytes exposed to 139 compounds for 24h and the highest dosage were used. This dataset was obtained from the OpenRiskNet service "Transcriptomics data from human, mouse, rat in vitro liver models”.
For the 139 compounds PubChem was used to obtain 2D Tanimoto scores and PIDGIN3 was used to retrieve ligand-based target predictions.
Integration of the transcriptomics data with the chemoinformatics data was performed using iClusterPlus, an integrative clustering framework developed to integrate diverse data types (i.e. binary, categorical, and continuous values) by a latent variable approach.
Technical implementation
Integration with other case studies is needed. SysGroup acquires information and data from the DataCure or TGX case studies, and can feed into AOPLink and ModelRX.
Through the services ToxPlanet and ToxicoDB of the Implementation Challenge winners Toxplanet and UHH, respectively, information on the obtained groups of chemicals is obtained.
Outcomes
The outcome of this case study is the workflow as shown in the figure above. This workflow was setup using GNU Make and is available through OpenRiskNet GitHub.
The workflow contains several steps to integrate three different datasets or data types for the purpose of grouping chemicals based on similarity in mechanisms of action (omics-based) and their chemoinformatics properties:
- Step 1: Obtaining transcriptomics data
- Step 2: Calculating a Tanimoto score for each compound
- Step 3: Predicting ligand-based protein targets for each compound
- Step 4: Grouping of chemicals from integrated data
Details on the implementation are included in the case study report linked below.
Currently available services:
-
Our platform searches the content from 500+ websites and quickly delivers relevant chemical hazard and toxicology data.Service type: Database / data source, Data mining tool, Service
-
Service type: Database / data source
-
A database for curated toxicogenomic datasetsService type: Database / data source, Application, Visualisation tool, Software
-
Interactive computing and workflows sharingService type: Visualisation tool, Helper tool, Software, Analysis tool, Processing tool, Workflow tool
-
Computation research made simple and reproducibleService type: Database / data source, Visualisation tool, Software, Analysis tool, Service, Workflow tool
Related resources
The OpenRiskNet case studies (originally outlined in Deliverable 1.3) were developed to demonstrate the modularised application of interoperable and interlinked workflows. These workflows were designed to address specific aspects required to inform on the potential of a compound to be toxic to humans and to eventually perform a risk assessment analysis. While each case study targets a specific area including data collection, kinetics modelling, omics data and Quantitative Structure Activity Relationships (QSAR), together they address a more complete risk assessment framework. Additionally, the modules here are fine-tuned for the utilisation and application of new approach methodologies (NAMs) in order to accelerate the replacement of animals in risk assessment scenarios. These case studies guided the selection of data sources and tools for integration and acted as examples to demonstrate the OpenRiskNet achievements to improve the level of the corresponding APIs with respect to harmonisation of the API endpoints, service description and semantic annotation.
This case study will use the approach of the diXa / DECO2 (Cefic-LRI AIMT4) projects to reproduce and extend the results obtained on the identification of hepatotoxicant groups based on similarity in mechanisms of action (omics-based) and chemical structure using services from OpenRiskNet.
Report
The OpenRiskNet project (https://openrisknet.org/) is funded by the H2020-EINFRA-22-2016 Programme. Here we present how the concept of Adverse Outcome Pathways (AOPs), which captures mechanistic knowledge from a chemical exposure causing a Molecular Initiating Event (MIE), through Key Events (KEs) towards an Adverse Outcome (AO), can be extended with additional knowledge by using tools and data available through the OpenRiskNet e-Infrastructure. This poster describes how the case study of AOPLink, together with DataCure, TGX, and SysGroup, can utilize the AOP framework for knowledge and data integration to support risk assessments. AOPLink involves the integration of knowledge captured in AOPs with additional data sources and experimental data from DataCure. TGX feeds this integration with prediction models of the MIE of such AOPs using either gene expression data or knowledge about stress response pathways. This is complemented by SysGroup, which is about the grouping of chemical compounds based on structural similarity and mode of action based on omics data. Therefore, the combination of these case studies extends the AOP knowledge and allows biological pathway analysis in the context of AOPs, by combining experimental data and the molecular knowledge that is captured in KEs of AOPs.
ToxTargetLinks