Case Study
AOPLink – Identification and Linking of Data related to AOPWiki
Summary
The Adverse Outcome Pathway (AOP) concept has been introduced to support risk assessment (Ankley et al., 2010). An AOP is initiated upon exposure to a stressor that causes a Molecular Initiating Event (MIE), followed by a series of Key Events (KEs) on increasing levels of biological organization. Eventually, the chain of KEs ends with the Adverse Outcome (AO), which describes the phenotypic outcome, disease, or the effect on the population.
In general, an AOP captures mechanistic knowledge of a sequence of toxicological responses after exposure to a stressor. While starting with molecular information, for example, the initial interaction of a chemical with a cell, the AOPs contain information of downstream responses of the tissue, organ, individual and population. Currently, AOPs are stored in the AOPWiki, a collaborative platform to exchange mechanistic toxicological knowledge as a part of the AOP-KB, an initiative by the OECD.
Normally, AOP development starts with a thorough literature search for existing knowledge, describing the sequence of KEs that form the AOP. However, the use of AOPs for regulatory purposes also requires detailed validation and linking to existing knowledge (Knapen et al., 2015; Burgdorf et al., 2017). Part of the development of AOPs is the search for data that supports the occurrence and biological plausibility of KEs and their relationships (KERs). This type of data can be found in literature, and increasingly in public databases.
The main goal of this case study is to establish the links between AOPs of the AOP-Wiki and experimental data to support a particular AOP. This will allow finding AOPs related to experimental data, and finding data related to a particular AOP.
Objectives
The objective of this case study is to establish how existing AOPs on AOP-Wiki can be linked to experimental bioassay data. The approach here is to link assay data via assay types to key events (KEs) in the AOP.
For this case study we aim to develop:
- FAIR (Findable, Accessible, Interoperable and Reusable) version of AOPWiki;
- Identifier mappings for MIEs, KEs, and biological and chemical entities (genes, proteins, metabolites);
- Establish links between MIEs and KEs to biological assays and experimental data;
- Establish links between assays and biological and chemical entities;
- Establish interoperable databases.
Risk assessment framework
The AOPLink case study covers a range of steps across different tiers of the SEURAT-1 risk assessment framework (Berggren et al., 2017). AOPLink allows finding relevant experimental data for given compounds and nanomaterials and KEs (Tier 0, step 3), identify biological processes affected by exposure to those chemicals supporting hypothesis generation (Tier 1, step 6), and using these sources of information to determine if an AOP can be applied to that chemical and if not what information is missing (Tier 3, step 9).
Link to other case studies
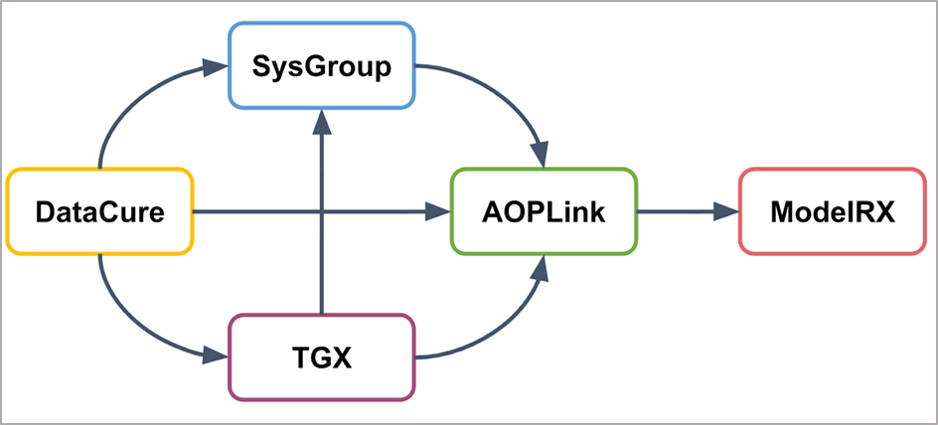
With respect to the other OpenRiskNet case studies, AOPLink has a strong link to Datacure, as the primary goal of AOPLink is the search for experimental datasets related to AOPs of interest. Furthermore, AOPLink can take as input from SysGroup on similar chemicals (same group) in case no direct search results are found with the chemical of interest. Also, TGX may provide predicted data to complement experimental data, to support searching, and predicting the activation of a range of Molecular Initiating Events (MIEs). Because AOPLink may result in hypothesis and list KERs, these results can be passed to ModelRX for further prediction and read-across.
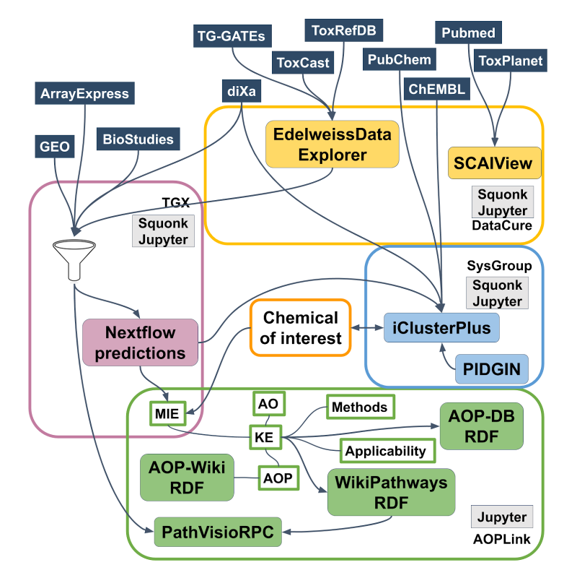
Databases and tools
The following sets of repositories and services are used in the AOPLink case study:
- AOP-related repositories: AOP-Wiki, AOP-DB;
- Biological pathway databases/tools: WikiPathways, Reactome;
- Experimental data repositories: diXa data warehouse, BioStudies ArrayExpress, ToxCast, ToxRefDB, TG-GATES, eNanoMapper, EPA Chemistry Dashboard, NORMAN Network;
- Identifier mapping services: BridgeDb, ChemIdConverter;
- Pathway analysis: PathVisioRPC;
Service integration
Services integrated for AOPLink
AOP-Wiki: The AOP-Wiki repository is part of the AOP Knowledge Base (AOP-KB), a joint effort of the US-Environmental Protection Agency and European Commission - Joint Research Centre. It is developed to facilitate collaborative AOP development, storage of AOPs, and therefore allow reusing toxicological knowledge for risk assessors. This Case Study has converted the AOP-Wiki XML data into an RDF schema, which has been exposed in a public SPARQL endpoint in the OpenRiskNet e-infrastructure.
EPA AOP Database (AOP-DB): The EPA AOP-DB supports the discovery and development of putative and potential AOPs. Based on public annotations, it integrates AOPs with gene targets, chemicals, diseases, tissues, pathways, species orthology information, ontologies, and gene interactions. The AOP-DB facilitates the translation of AOP biological context, and associates assay, chemical and disease endpoints with AOPs (Pittman et al., 2018; Mortensen et al., 2018). The AOP-DB won the first OpenRiskNet implementation challenge of the associated partner program and is therefore integrated into the OpenRiskNet e-infrastructure. After the conversion of the AOP-DB into an RDF schema, its data will be exposed in a Virtuoso SPARQL endpoint.
WikiPathways and Reactome: WikiPathways is a community-driven molecular pathway database, supporting wide-spread topics and supported by many databases and integrative resources. It contains semantic annotations in its pathways for genes, proteins, metabolites, and interactions using a variety of reference databases, and WikiPathways is used to analyze and integrate experimental omics datasets (Slenter et al., 2017). Furthermore, human pathways from Reactome (Fabregat et al., 2018), another molecular pathway database, are integrated with WikiPathways and are therefore part of the WikiPathways RDF (Waagmeester et al., 2016). On the OpenRiskNet e-infrastructure, the WikiPathways RDF, which includes the Reactome pathways, is exposed via a Virtuoso SPARQL endpoint.
eNanoMapper: The eNanoMapper database hosts nanomaterials characterization data and biological and toxicological information. It allows users to upload and explore data and information about nanomaterials through a REST web services API and a web browser interface, which is available in the OpenRiskNet e-infrastructure, using a newly developed Docker image.
BridgeDb: In order to link databases and services that use particular identifiers for genes, proteins, and chemicals, the BridgeDb platform is integrated into the OpenRiskNet e-infrastructure. It allows for identifier mapping between various biological databases for data integration and interoperability (van Iersel et al., 2010).
PathVisioRPC: To allow the analysis and visualization of transcriptomics or metabolomics data, PathVisioRPC (Bohler et al., 2015) will be used in AOPLink workflows. It is an XML-RPC interface, available for use in a variety of coding environments. It supports the use of pathways from WikiPathways for pathway statistics, exporting of results and providing data visualization on the pathways.
Services provided by other case studies
- EdelweissData: Curated datasets are made available through the EdelweissData Explorer, the main data provisioning tool in the DataCure case study. It is a web-based data explorer tool that gives users the ability to filter, search and extract data through the use of API calls. The EdelweissData Explorer serves data from ToxCast, ToxRefDB, and TG-GATES.
- ChemIdConverter: The ChemIdConverter allows users to submit and translate a variety of chemical descriptors, such as SMILES and InChI, through a REST API.
- Grouping service that classifies a chemical or nanomaterial and provides structurally and/or biologically similar chemical substances or compounds.
TGX:
- API to access predicted data for the activation of a selective set of Molecular Initiating Events.
Technical implementation
Jupyter Notebooks were used to integrate the tools into reproducible workflows. Of the services, data from the AOP-Wiki, AOP-DB, and WikiPathways are available through SPARQL endpoints and are easily queried with the SPARQLwrapper python library. The other services, which have OpenAPI 2 or 3 definitions, are called through direct API calls. Combining the various tools and databases into workflows, the research goals are answered with modular, reproducible Jupyter Notebooks. For example, workflows are developed to find public experimental data that supports AOPs and to retrieve AOPs that are related to available data. Furthermore, we aim to develop complete data analysis workflows using WikiPathways and PathVisioRPC and relate the results to the knowledge captured in AOP-Wiki and AOP-DB.
Outcomes
This case study focused mainly on the use of AOP knowledge, and extend it with additional information, or experimental data. The main repository in those analyses was the AOP-Wiki, which was converted into RDF, deployed in a Virtuoso SPARQL endpoint.
AOPs linked to WikiPathways
One of the first analyses in the AOPLink case study was the assessment of the possible links between AOPs of AOP-Wiki, and the molecular pathways of WikiPathways. This task involved the identification of ontology usage for describing biological processes, and looking for the overlap of chemical coverage in both repositories. This exercise showed that few of the AOP-linked chemicals are found in WikiPathways, whereas 70% of all mapped genes are present in molecular pathways on WikiPathways. A manual assessment indicated that 67% of all low-level KEs, including molecular, cellular, tissue and organ KEs, can be linked (partially) to molecular pathways
(Martens et al., 2018).
Workflow for finding data related to an AOP
One of the main questions to solve in AOPLink is the finding of data that supports an AOP of interest. To answer that, we have developed a Jupyter notebook that does that by using the AOP-Wiki RDF, AOP-DB RDF, BridgeDb, EdelweissData explorer, and WikiPathways services. The workflow, which was also presented during the workflow tutorial at the final workshop of OpenRiskNet, AOP 37 was selected as the AOP of interest.
First, the AOP-Wiki RDF was used to extract information about the AOP by using a variety of SPARQL queries that directly access the data through the SPARQL endpoint with the SPARQLWrapper library. Information of the AOP, such as the title, abstract, KEs and stressors, were extracted and written in a data frame, along with an AOP network that displays the connected AOPs.
Additional details on the outcomes are included in the case study report linked below.
Currently available services:
-
Service type: Database / data source
-
Service type: Database / data source, Service
-
ToxCast and Tox21 datasets (raw and summary) extracted from the MySQL database provided by US EPAService type: Database / data source
-
Service type: Database / data source
-
Service type: Database / data source
-
Service type: Database / data source
-
ZAPService type: Database / data source
-
Extend molecular biological networks with toxic compounds.Service type: Database / data source
-
Service type: Database / data source
-
Toxicology database, Knowledge base and data mining, AOP related toolsService type: Database / data source, Service
-
Processed data (counts and fold changes) based on the transcriptomics data provided by Open TG-GATEs and DrugMatrixService type: Database / data source
-
Interactive computing and workflows sharingService type: Visualisation tool, Helper tool, Software, Analysis tool, Processing tool, Workflow tool
Related resources
The OpenRiskNet case studies (originally outlined in Deliverable 1.3) were developed to demonstrate the modularised application of interoperable and interlinked workflows. These workflows were designed to address specific aspects required to inform on the potential of a compound to be toxic to humans and to eventually perform a risk assessment analysis. While each case study targets a specific area including data collection, kinetics modelling, omics data and Quantitative Structure Activity Relationships (QSAR), together they address a more complete risk assessment framework. Additionally, the modules here are fine-tuned for the utilisation and application of new approach methodologies (NAMs) in order to accelerate the replacement of animals in risk assessment scenarios. These case studies guided the selection of data sources and tools for integration and acted as examples to demonstrate the OpenRiskNet achievements to improve the level of the corresponding APIs with respect to harmonisation of the API endpoints, service description and semantic annotation.
The Adverse Outcome Pathway (AOP) concept has been introduced to support risk assessment (Ankley et al., 2010). An AOP is initiated upon exposure to a stressor that causes a Molecular Initiating Event (MIE), followed by a series of Key Events (KEs) on increasing levels of biological organization. Eventually, the chain of KEs ends with the Adverse Outcome (AO), which describes the phenotypic outcome, disease, or the effect on the population. In general, an AOP captures mechanistic knowledge of a sequence of toxicological responses after exposure to a stressor. While starting with molecular information, for example, the initial interaction of a chemical with a cell, the AOPs contain information of downstream responses of the tissue, organ, individual and population. Currently, AOPs are stored in the AOP-Wiki, a collaborative platform to exchange mechanistic toxicological knowledge as a part of the AOP-KB, an initiative by the OECD. Normally, AOP development starts with a thorough literature search for existing knowledge, describing the sequence of KEs that form the AOP. However, the use of AOPs for regulatory purposes also requires detailed validation and linking to existing knowledge (Knapen et al., 2015; Burgdorf et al., 2017). Part of the development of AOPs is the search for data that supports the occurrence and biological plausibility of KEs and their relationships (KERs). This type of data can be found in literature, and increasingly in public databases. The main goal of this case study is to establish the links between AOPs of the AOP-Wiki and experimental data to support a particular AOP. This will allow finding AOPs related to experimental data, and finding data related to a particular AOP.
Report
The OpenRiskNet project (https://openrisknet.org/) is funded by the H2020-EINFRA-22-2016 Programme. Here we present how the concept of Adverse Outcome Pathways (AOPs), which captures mechanistic knowledge from a chemical exposure causing a Molecular Initiating Event (MIE), through Key Events (KEs) towards an Adverse Outcome (AO), can be extended with additional knowledge by using tools and data available through the OpenRiskNet e-Infrastructure. This poster describes how the case study of AOPLink, together with DataCure, TGX, and SysGroup, can utilize the AOP framework for knowledge and data integration to support risk assessments. AOPLink involves the integration of knowledge captured in AOPs with additional data sources and experimental data from DataCure. TGX feeds this integration with prediction models of the MIE of such AOPs using either gene expression data or knowledge about stress response pathways. This is complemented by SysGroup, which is about the grouping of chemical compounds based on structural similarity and mode of action based on omics data. Therefore, the combination of these case studies extends the AOP knowledge and allows biological pathway analysis in the context of AOPs, by combining experimental data and the molecular knowledge that is captured in KEs of AOPs.
ToxTargetLinks
BridgeDb identifier mapping service
AOP-Wiki SPARQL Endpoint
ToxTargetLinks
The Adverse Outcome Pathway Database (AOP-DB)
Slides
BridgeDb identifier mapping service
AOP-Wiki SPARQL Endpoint
ToxTargetLinks
The Adverse Outcome Pathway Database (AOP-DB)
Slides
Slides
Slides
BridgeDb identifier mapping service
AOP-DB SPARQL Endpoint
AOP-Wiki SPARQL Endpoint
ToxTargetLinks
1. Introduction With the ever-growing number of chemicals that require toxicological risk assessment, there is a need for faster, more efficient use of existing data to assemble effective assessment strategies [1]. Therefore, the concept of Adverse Outcome Pathways (AOPs) was introduced [2], a framework to organize existing mechanistic information about toxicological processes into a chain of smaller pieces of knowledge, called Key Events (KEs). These allow the structuring of toxicological knowledge and reduce the effort needed to capture all information before performing risk assessment [2, 3]. In order to facilitate a community effort in gathering toxicological knowledge, the AOP-Wiki was created by the European Commission JRC and the US EPA. To integrate this knowledge base more easily with other resources, we explored the use of semantic web technologies to link AOP-Wiki with other chemical and biological databases. 2. Approach The AOP-Wiki provides quarterly permanent downloads for the full database XML (https://aopwiki.org/downloads/). We parsed the AOP-Wiki knowledge with Python 3.5 and the ElementTree XML API and converted it into a semantic web RDF format, which allows for accurate description with ontological annotations, including the AOPO, CHEMINF, and Dublin Core. Chemical compounds are identified in the AOP-Wiki with CAS numbers and biological processes with a variety of ontologies, e.g. GO, Mammalian Phenotype Ontology, and Molecular Interactions ontology. These annotations are used to create Internationalized Resource Identifiers. To integrate and test the RDF, a variety of federated SPARQL queries were written and executed in Blazegraph (build version 2.1.4). 3. Results We created an AOP-Wiki RDF scheme and converted the XML into Turtle syntax. The RDF was tested with a variety of SPARQL queries to answer biological question relevant to risk assessment, such as: - What measurement / test-method information is available for a given AOP? - Which of the stressor chemicals on the AOP-Wiki can be linked molecular pathways on WikiPathways? 4. Discussion The RDF transformation of AOP-Wiki content can assist in the accessibility and expansion of toxicological knowledge by allowing semantic interoperability. The created RDF of the AOP-Wiki allows the querying and providing of additional information for stressor chemicals, genes, and proteins involved in KEs, the underlying molecular pathways, but also for the applicability of AOPs by cell types or species. This semantic approach allows novel ways to explore the rapidly growing AOP knowledge with every new publication related to toxicological studies. There is work in progress on a Virtuoso SPARQL endpoint Docker image to simplify the use of the data, and integrate the database in the OpenRiskNet e-infrastructure to provide AOP knowledge useful for automated risk assessment workflows. Funding This project has received funding from the European Union’s Horizon 2020 (EU 2020) research and innovation program under grant agreement no. 681002 (EU-ToxRisk) and EINFRA-22-2016 program under grant agreement no. 731075 (OpenRiskNet).
A paradigm shift is taking place in risk assessment to replace animal models, reduce the number of economic resources, and refine the methodologies to test the growing number of chemicals and nanomaterials. Therefore, approaches such as transcriptomics, proteomics, and metabolomics have become valuable tools in toxicological research, and are finding their way into regulatory toxicity. One promising framework to bridge the gap between the molecular-level measurements and risk assessment is the concept of Adverse Outcome Pathways (AOPs). These pathways comprise mechanistic knowledge and connect biological events from a molecular level towards an adverse effect outcome after exposure to a chemical. However, the implementation of omics-based approaches in the AOPs and their acceptance by the risk assessment community is still a challenge.
Example workflow based on OpenRiskNet tools - Pathway identification workflow related to DataCure and AOPlink case studies. This notebook downloads TG-Gates data of 4 compounds and selects genes overexpressed in all sample. The Affymetrix probe sets are then translated into Ensembl gene identifiers using the BridgeDB service and pathways associated with the genes are identified using the WikiPathways service.
BridgeDb identifier mapping service
EdelweissData serving ToxCast, ToxRefDB and TG-GATEs data
Jupyter Notebooks